Characteristics of root-associated microbiomes and their responses to soil nitrogen levels in different wheat cultivars
-
摘要: 植物根区微生物在植物养分吸收以及生长发育过程中发挥重要作用。为研究不同小麦品种根区微生物群落结构差异, 本研究选择‘科农9204’ ‘科农2011’ ‘京411’和‘百农207’ 4个小麦品种, 于高氮[300 kg(N)·hm−2]和低氮[0 kg(N)·hm−2]条件下进行田间试验, 在分蘖期、拔节期和灌浆期采集根区样品。通过16S rRNA基因高通量测序技术比较分析不同小麦品种根际土壤和根内生细菌群落结构和多样性, 同时测定小麦植株的生理参数。结果表明, ‘科农9204’在3个生育期及不同氮素水平下相对其他3个品种均具有较高的地上部氮素吸收量(除分蘖期低氮条件)。小麦根际土壤和根内生细菌群落的优势菌门均为变形菌门(Proteobacteria)和放线菌门(Actinobacteria)。相比其他3个品种, ‘科农 9204’ 的根际土壤细菌群落在拔节期低氮水平下富集了根瘤菌目(Rhizobiales)和芽单胞菌属(Gemmatimonas), 在灌浆期高氮水平下富集了弗兰克氏菌目(Frankiales)。相关性分析表明, 根际土壤细菌群落中的节杆菌属(Arthrobacter)、链霉菌属(Streptomyces)、红色杆菌属(Rubrobacter)和类诺卡氏菌属(Nocardioides)与小麦地上部生物量和地上部氮素吸收量呈显著正相关, 马赛菌属(Massilia)、砂单胞菌属(Arenimonas)、假单胞菌属(Pseudomonas)和黄杆菌属(Flavobacterium)与小麦地上部全氮含量呈显著正相关。上述结果表明, 小麦可能通过调控根区微生物群落来影响养分吸收, 且这种影响具有品种特异性。本研究为明确小麦与微生物互作机制、发掘对小麦有益的微生物并应用到农业生产中提供了依据。Abstract: Plant root-associated microorganisms play important roles in nutrient uptake and plant growth. In order to illustrate the differences in the root-associated microbial community structure of different wheat cultivars, four wheat cultivars (i.e., ‘Kenong 9204’ ‘Kenong 2011’ ‘Jing 411’ and ‘Bainong 207’) were planted under 0 kg(N)·hm−2 (low nitrogen level) and 300 kg(N)·hm−2 (high nitrogen level), and the rhizosphere and root samples were collected at the tillering, jointing, and filling stages. The bacterial diversity and community structure in the rhizosphere and root endosphere of different wheat cultivars were analyzed using 16S rRNA high-throughput sequencing, and the physiological parameters of wheat were determined. Compared with the other three cultivars, ‘Ke-nong 9204’ had higher aboveground nitrogen accumulation at the three growth stages and under two nitrogen levels, except at the tillering stage with a low nitrogen level. Proteobacteria and Actinobacteria were the dominant bacteria in the wheat rhizosphere and root endosphere. Compared to the other three cultivars, ‘Kenong 9204’ enriched Rhizobiales and Gemmatimonas in the rhizosphere soil bacterial community under low nitrogen level at the jointing stage and enriched Frankiales under high nitrogen level at the filling stage. Correlation analysis showed that Arthrobacter, Streptomyces, Rubrobacter, and Nocardioides in the rhizosphere soil bacterial communities were significantly positively correlated with aboveground biomass and nitrogen accumulation; Massilia, Arenimonas, Pseudomonas, and Flavobacterium were significantly positively correlated with aboveground nitrogen content. Our results indicate that wheat may affect nutrient uptake by regulating the composition of the microbial community in the root zone and that this effect is cultivar-specific. This study provides useful information for understanding plant-microbe interactions in wheat and harnessing beneficial microbes for agricultural production.
-
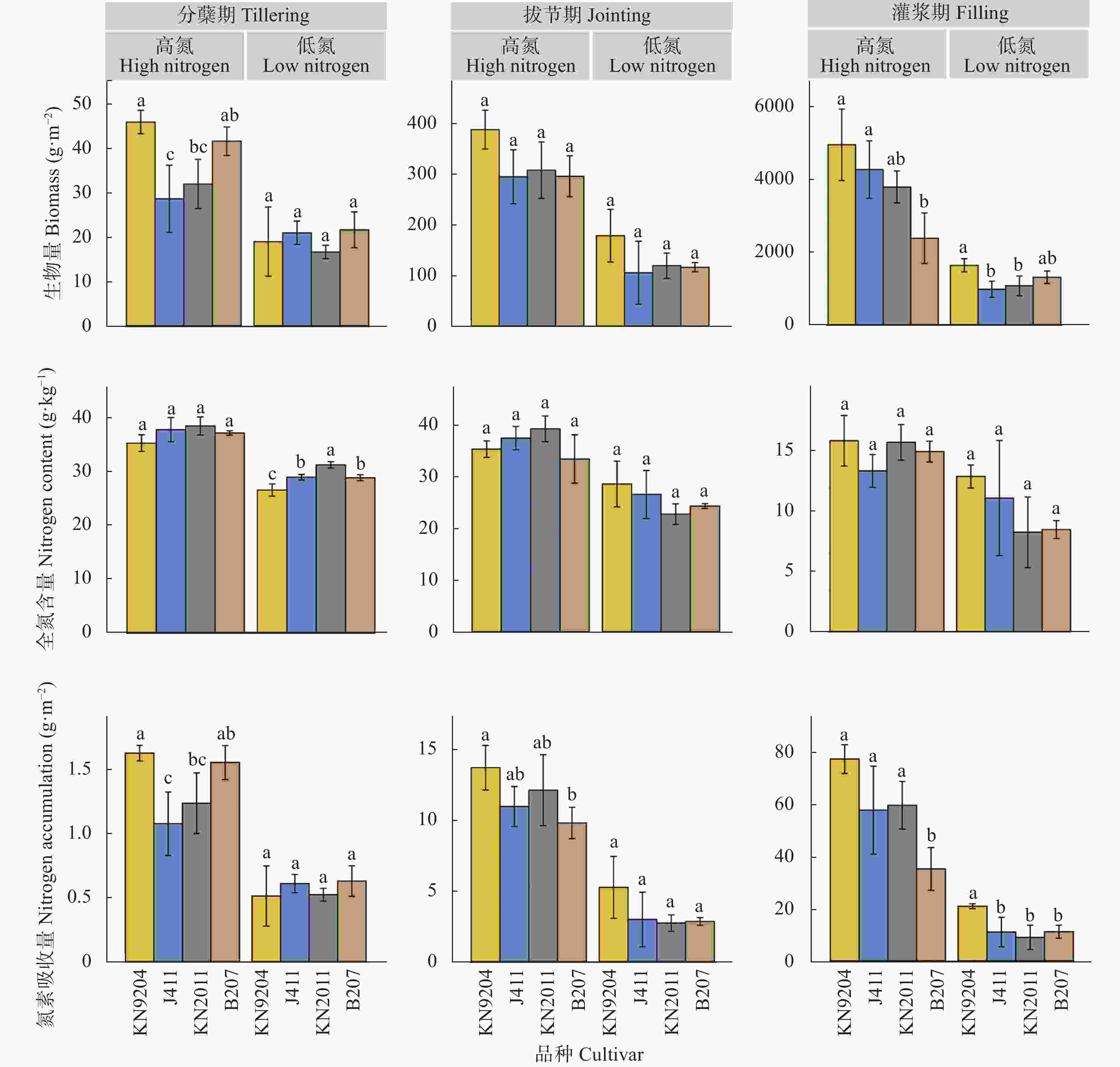
图 1 不同小麦品种地上部性状对比分析
不同小写字母代表不同小麦品种间差异显著(P<0.05)。KN9204、KN2011、J411和B207分别为小麦品种‘科农9204’ ‘科农2011’ ‘京411’和‘百农207’。Different lowercase letters represent significant differences among different wheat cultivars (P<0.05). KN9204, KN2011, J411 and B207 represent wheat cultivars of ‘Kenong 9204’ ‘Kenong 2011’ ‘Jing 411’ and ‘Bainong 207’, respectively.
Figure 1. Comparative analysis of aboveground part traits of different wheat cultivars
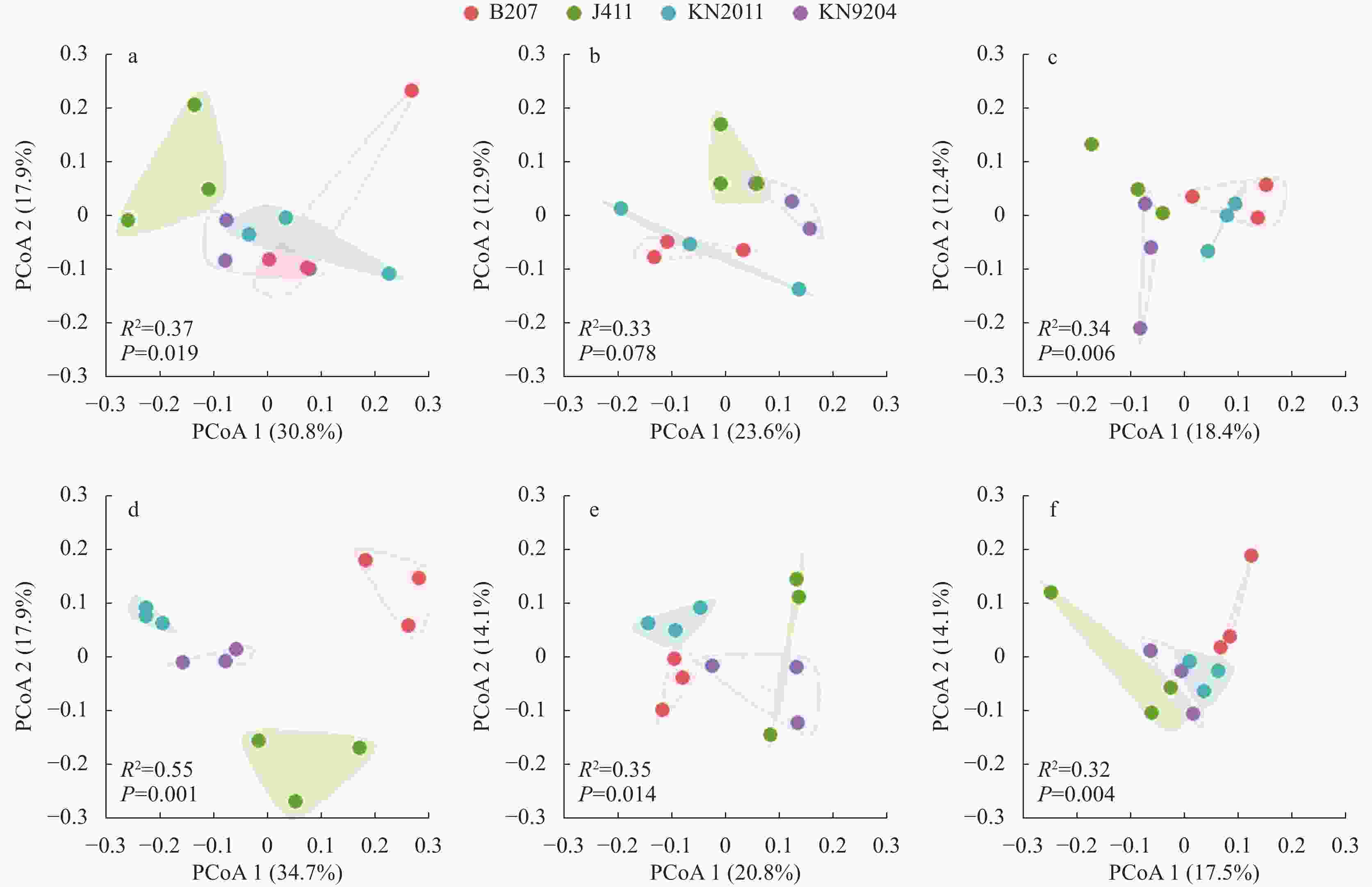
图 2 不同施氮水平下不同小麦品种根际土壤细菌群落的主坐标分析(PCoA)
a: 分蘖期-高氮; b: 拔节期-高氮; c: 灌浆期-高氮; d: 分蘖期-低氮; e: 拔节期-低氮; f: 灌浆期-低氮。KN9204、KN2011、J411和B207分别为小麦品种‘科农9204’ ‘科农2011’ ‘京411’和‘百农207’。PCoA1和PCoA2是两个主坐标成分。a: tillering stage at high nitrogen level; b: jointing stage at high nitrogen level; c: filling stage at high nitrogen level; d: tillering stage at low nitrogen level; e: jointing stage at low nitrogen level; f: filling stage at low nitrogen level. KN9204, KN2011, J411 and B207 represent wheat cultivars of ‘Kenong 9204’ ‘Kenong 2011’ ‘Jing 411’ and ‘Bainong 207’, respectively. PCoA1 and PCoA2 are the two principal coordinate components.
Figure 2. Principal coordinate analysis (PCoA) of rhizosphere soil bacterial communities of different wheat cultivars at different nitrogen levels
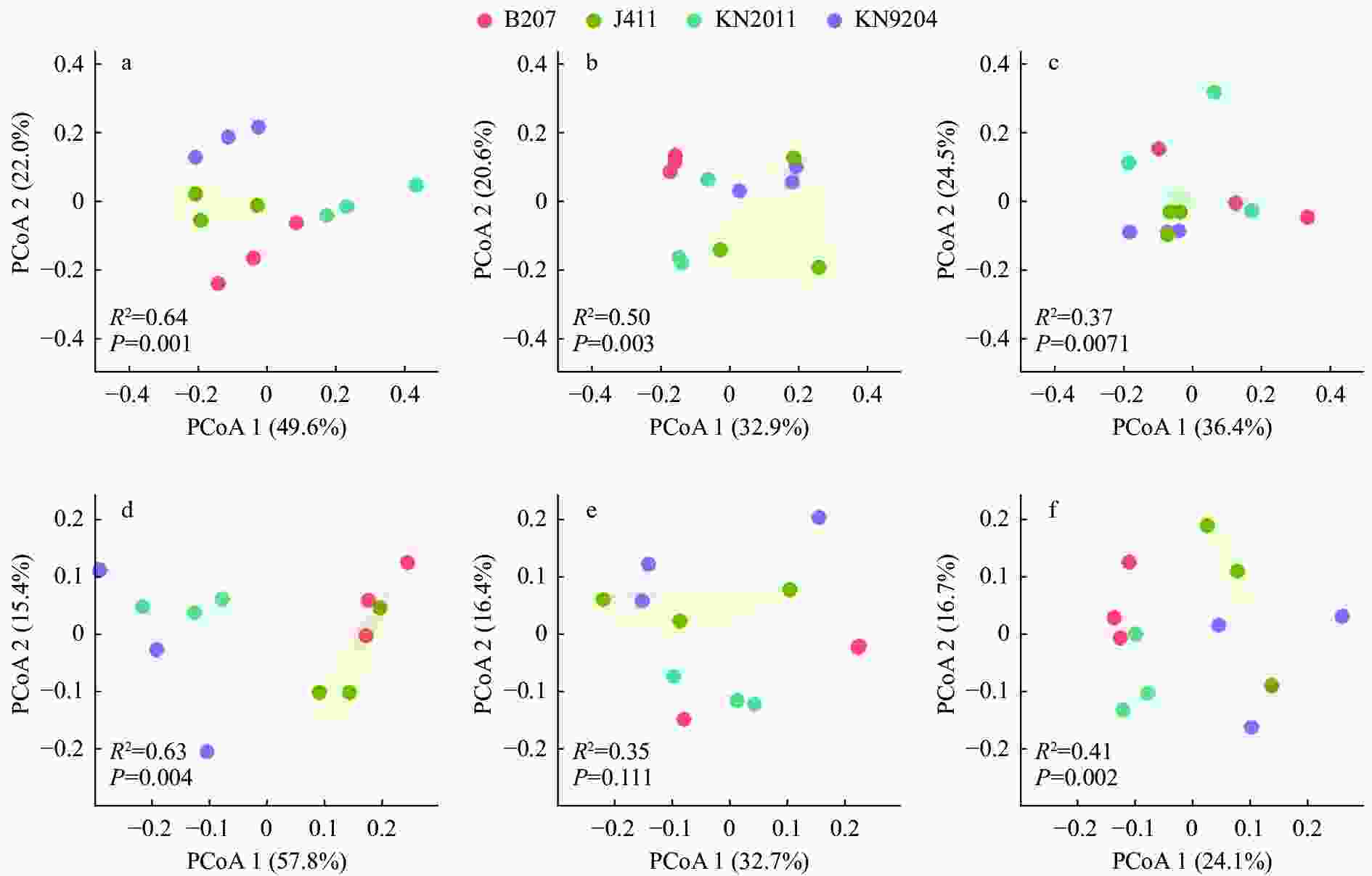
图 3 不同施氮水平下不同小麦品种根内生细菌群落的主坐标分析(PCoA)
a: 分蘖期-高氮; b: 拔节期-高氮; c: 灌浆期-高氮; d: 分蘖期-低氮; e: 拔节期-低氮; f: 灌浆期-低氮。KN9204、KN2011、J411和B207分别为小麦品种‘科农9204’ ‘科农2011’ ‘京411’和‘百农207’。PCoA1和PCoA2是两个主坐标成分。a: tillering stage at high nitrogen level; b: jointing stage at high nitrogen level; c: filling stage at high nitrogen level; d: tillering stage at low nitrogen level; e: jointing stage at low nitrogen level; f: filling stage at low nitrogen level. KN9204, KN2011, J411 and B207 represent wheat cultivars of ‘Kenong 9204’ ‘Kenong 2011’ ‘Jing 411’ and ‘Bainong 207’, respectively. PCoA1 and PCoA2 are the two principal coordinate components.
Figure 3. Principal coordinate analysis (PCoA) of root endosphere bacterial communities of different wheat cultivars at different nitrogen levels
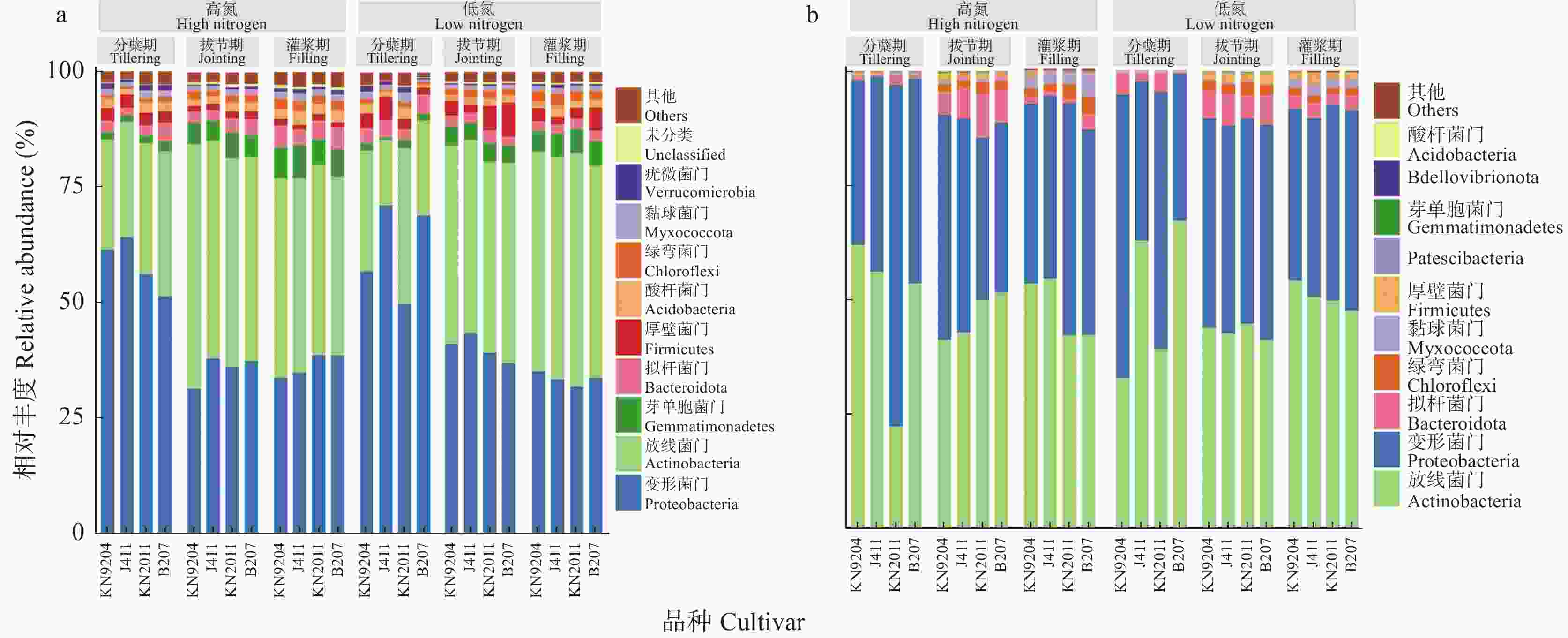
图 4 不同施氮水平下不同小麦品种根际土壤细菌(a)和根内生细菌(b)群落门水平相对丰度
KN9204、KN2011、J411和B207分别为小麦品种‘科农9204’ ‘科农2011’ ‘京411’和‘百农207’。KN9204, KN2011, J411 and B207 represent wheat cultivars of ‘Kenong 9204’ ‘Kenong 2011’ ‘Jing 411’ and ‘Bainong 207’, respectively.
Figure 4. Relative abundance of rhizosphere soil (a) and root endosphere (b) bacterial community at the phylum level of different wheat cultivars at different nitrogen levels
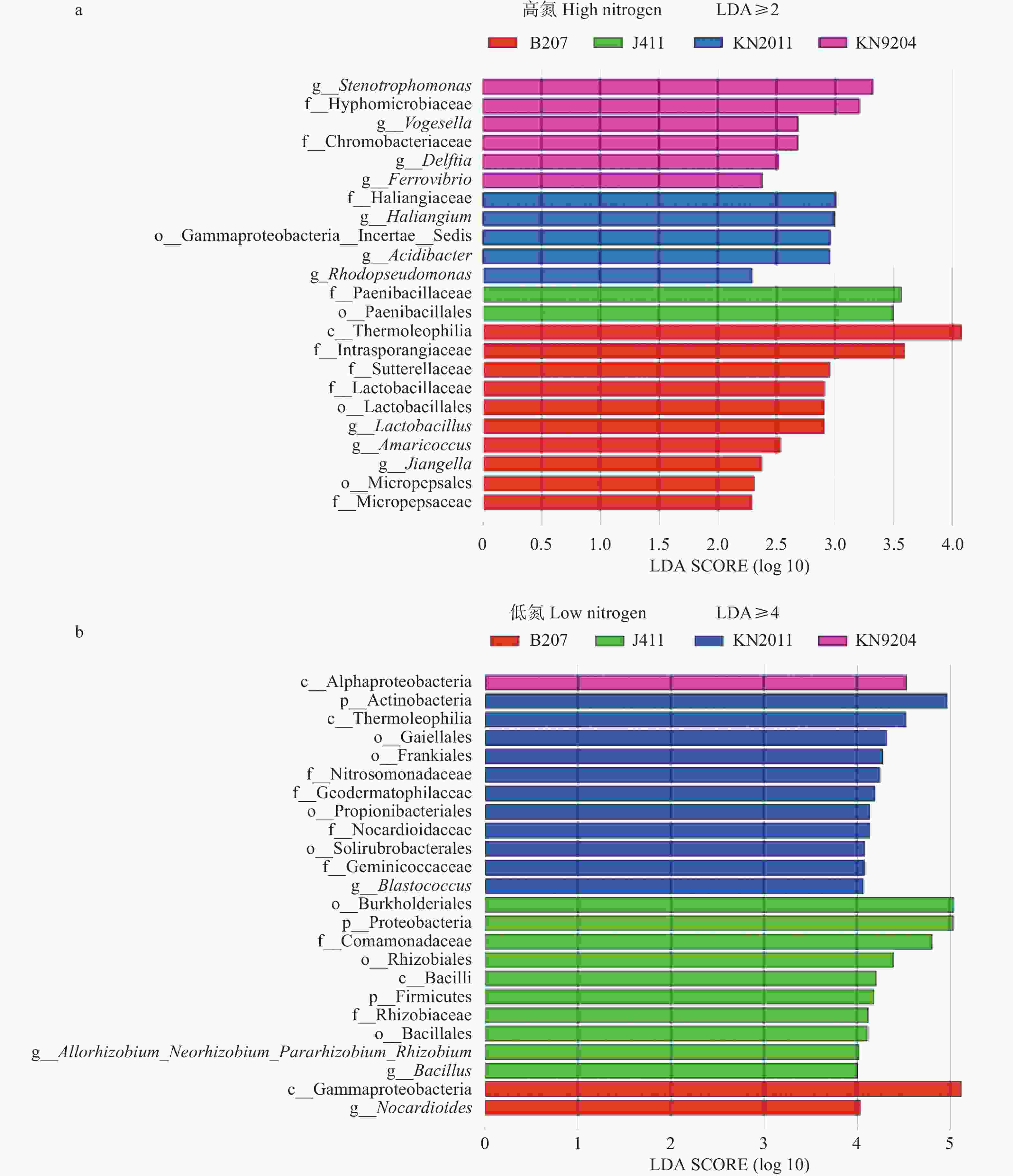
图 5 不同施氮水平下分蘖期不同小麦品种根际土壤微生物差异分析
纵坐标中的字母p、c、o、f和g为分类水平, 分别代表门、纲、目、科和属。KN9204、KN2011、J411和B207分别为小麦品种‘科农9204’ ‘科农2011’ ‘京411’和‘百农207’。LDA SCORE表示差异显著物种的影响力。 p, c, o, f and g in the labels for vertical axis represent classification level, representing phylum, class, order, family and genus, respectively. KN9204, KN2011, J411 and B207 represent wheat cultivars of ‘Kenong 9204’ ‘Kenong 2011’ ‘Jing 411’ and ‘Bainong 207’, respectively. LDA SCORE refers to the effect size of significantly different species.
Figure 5. Analysis on the difference in rhizosphere soil microorganisms among different wheat cultivars at tillering stage at different nitrogen levels
图 6 不同施氮水平下拔节期不同品种小麦根际土壤微生物差异分析
纵坐标中的字母p、c、o、f和g为分类水平, 分别代表门、纲、目、科和属。KN9204、KN2011、J411和B207分别为小麦品种‘科农9204’ ‘科农2011’ ‘京411’和‘百农207’。LDA SCORE表示差异显著物种的影响力。p, c, o, f and g in the labels for vertical axis represent classification level, representing phylum, class, order, family and genus, respectively. KN9204, KN2011, J411 and B207 represent wheat cultivars of ‘Kenong 9204’ ‘Kenong 2011’ ‘Jing 411’ and ‘Bainong 207’, respectively. LDA SCORE refers to the effect size of significantly different species.
Figure 6. Analysis on the difference in rhizosphere soil microorganisms among different wheat cultivars at jointing stage at different nitrogen levels
图 7 不同施氮水平下灌浆期不同品种小麦根际土壤微生物差异分析
纵坐标中的字母p、c、o、f和g为分类水平, 分别代表门、纲、目、科和属。KN9204、KN2011、J411和B207分别为小麦品种‘科农9204’ ‘科农2011’ ‘京411’和‘百农207’。LDA SCORE表示差异显著物种的影响力。p, c, o, f and g in the lables for vertical axisrepresent classification level, representing phylum, class, order, family and genus, respectively. KN9204, KN2011, J411 and B207 represent wheat cultivars of ‘Kenong 9204’ ‘Kenong 2011’ ‘Jing 411’ and ‘Bainong 207’, respectively. LDA SCORE refers to the effect size of significantly different species.
Figure 7. Analysis on the difference in rhizosphere soil microorganisms among different wheat cultivars at filling stage at different nitrogen levels
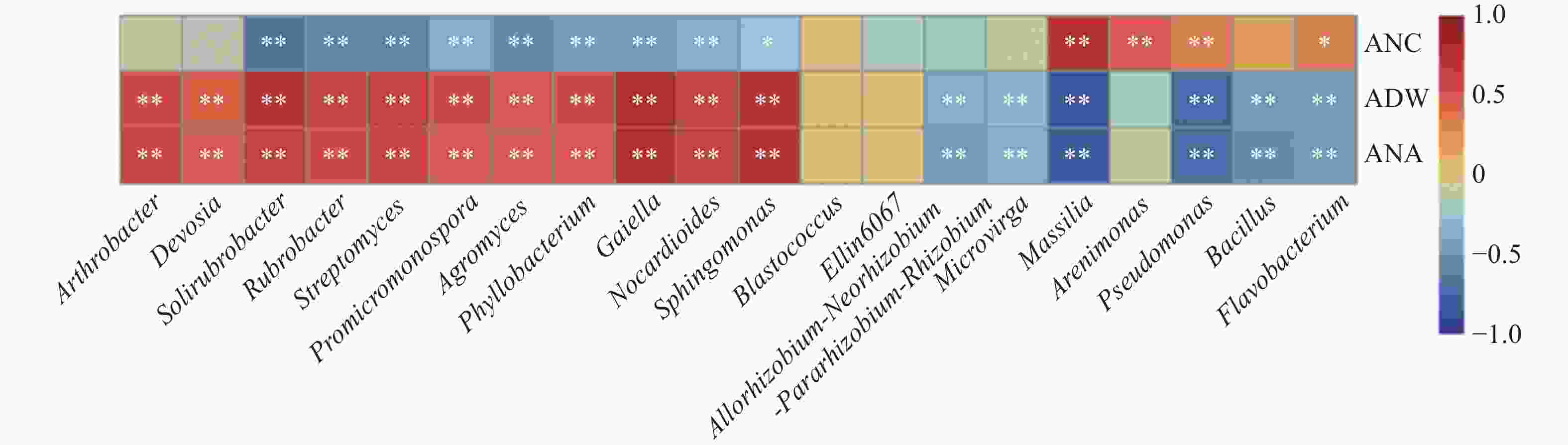
图 8 小麦根际土壤微生物与地上部性状的相关性分析
**和*分别表示在P<0.01和P<0.05水平显著相关。ANC、ADW和ANA分别为地上部全氮含量、地上部生物量和地上部氮素吸收量。** and * represent significant differences at P<0.01 and P<0.05 levels, respectively. ANC, ADW and ANA are nitrogen content, aboveground biomass and nitrogen accumulation of wheat, respectively.
Figure 8. Correlation analysis between rhizosphere soil microorganisms and traits of wheat aboveground part
表 1 不同品种小麦根际土壤和根内生细菌群落的Alpha多样性
Table 1. Alpha diversity of rhizosphere soil and endosphere bacterial community in different wheat cultivars
取样部位
Compartment施氮水平
Nitrogen level时期
Growth stage科农9204
Kenong 9204京411
Jing 411科农2011
Kenong 2011百农207
Bainong 207根际土壤
Rhizosphere soil高氮
High nitrogen分蘖期 Tillering 8.72±0.41a 7.49±0.79b 8.92±0.60a 9.18±0.46a 拔节期 Jointing 8.89±0.29a 8.97±0.03a 9.00±0.30a 9.18±0.10a 灌浆期 Filling 9.34±0.21a 9.47±0.06a 9.29±0.07a 9.25±0.14a 低氮
Low nitrogen分蘖期 Tillering 9.29±0.21a 8.56±0.55b 9.58±0.10a 7.85±0.17c 拔节期 Jointing 9.23±0.06a 9.06±0.09ab 8.93±0.06b 8.97±0.17b 灌浆期 Filling 9.29±0.1ab 9.36±0.05a 9.24±0.10ab 9.16±0.12b 根内
Root endosphere高氮
High nitrogen分蘖期 Tillering 5.56±0.18a 5.43±0.33a 5.48±0.22a 5.32±0.61a 拔节期 Jointing 6.79±0.20a 6.42±0.47a 6.84±0.14a 6.64±0.18a 灌浆期 Filling 6.55±0.38b 6.51±0.12b 6.96±0.55ab 7.36±0.39a 低氮
Low nitrogen分蘖期 Tillering 5.54±0.33a 4.58±0.6bc 5.19±0.08ab 4.09±0.68c 拔节期 Jointing 6.17±0.16a 6.53±0.56a 6.68±0.35a 6.81±0.35a 灌浆期 Filling 6.50±0.60a 6.89±0.06a 6.87±0.20a 7.10±0.06a 不同小写字母代表不同小麦品种间差异显著(P<0.05)。Different lowercase letters represent significant differences among different wheat cultivars (P<0.05). -
[1] MÜLLER D B, VOGEL C, BAI Y, et al. The plant microbiota: systems-level insights and perspectives[J]. Annual Review of Genetics, 2016, 50: 211−234 doi: 10.1146/annurev-genet-120215-034952 [2] FITZPATRICK C R, SALAS-GONZÁLEZ I, CONWAY J M, et al. The plant microbiome: from ecology to reductionism and beyond[J]. Annual Review of Microbiology, 2020, 74: 81−100 doi: 10.1146/annurev-micro-022620-014327 [3] BERENDSEN R L, PIETERSE C M J, BAKKER P A H M. The rhizosphere microbiome and plant health[J]. Trends in Plant Science, 2012, 17(8): 478−486 doi: 10.1016/j.tplants.2012.04.001 [4] VERBON E H, LIBERMAN L M. Beneficial microbes affect endogenous mechanisms controlling root development[J]. Trends in Plant Science, 2016, 21(3): 218−229 doi: 10.1016/j.tplants.2016.01.013 [5] SCHLEMPER T R, LEITE M F A, LUCHETA A R, et al. Rhizobacterial community structure differences among sorghum cultivars in different growth stages and soils[J]. FEMS Microbiology Ecology, 2017, 93(8): 1−11 [6] LI T, LI Y Z, GAO X C, et al. Rhizobacterial communities and crop development in response to long-term tillage practices in maize and soybean fields on the Loess Plateau of China[J]. Catena, 2021, 202: 105319 doi: 10.1016/j.catena.2021.105319 [7] YU P, HE X M, BAER M, et al. Plant flavones enrich rhizosphere Oxalobacteraceae to improve maize performance under nitrogen deprivation[J]. Nature Plants, 2021, 7(4): 481−499 doi: 10.1038/s41477-021-00897-y [8] KWAK M J, KONG H G, CHOI K, et al. Rhizosphere microbiome structure alters to enable wilt resistance in tomato[J]. Nature Biotechnology, 2018, 36(11): 1100−1109 doi: 10.1038/nbt.4232 [9] MENDES L W, RAAIJMAKERS J M, DE HOLLANDER M, et al. Influence of resistance breeding in common bean on rhizosphere microbiome composition and function[J]. The ISME Journal, 2018, 12(1): 212−224 doi: 10.1038/ismej.2017.158 [10] CHAI X, WANG L, YANG Y, et al. Apple rootstocks of different nitrogen tolerance affect the rhizosphere bacterial community composition[J]. Journal of Applied Microbiology, 2019, 126(2): 595−607 doi: 10.1111/jam.14121 [11] MAHONEY A K, YIN C T, HULBERT S H. Community structure, species variation, and potential functions of rhizosphere-associated bacteria of different winter wheat (Triticum aestivum) cultivars[J]. Frontiers in Plant Science, 2017, 8: 132 [12] KAVAMURA V N, ROBINSON R J, HUGHES D, et al. Wheat dwarfing influences selection of the rhizosphere microbiome[J]. Scientific Reports, 2020, 10(1): 1452 doi: 10.1038/s41598-020-58402-y [13] KRAISER T, GRAS D E, GUTIÉRREZ A G, et al. A holistic view of nitrogen acquisition in plants[J]. Journal of Experimental Botany, 2011, 62(4): 1455−1466 doi: 10.1093/jxb/erq425 [14] GUO J H, LIU X J, ZHANG Y, et al. Significant acidification in major Chinese croplands[J]. Science, 2010, 327(5968): 1008−1010 doi: 10.1126/science.1182570 [15] LIU X J, ZHANG F S. Nitrogen fertilizer induced greenhouse gas emissions in China[J]. Current Opinion in Environmental Sustainability, 2011, 3(5): 407−413 doi: 10.1016/j.cosust.2011.08.006 [16] CHEN S M, WAGHMODE T R, SUN R B, et al. Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization[J]. Microbiome, 2019, 7(1): 136 doi: 10.1186/s40168-019-0750-2 [17] ZHU S S, VIVANCO J M, MANTER D K. Nitrogen fertilizer rate affects root exudation, the rhizosphere microbiome and nitrogen-use-efficiency of maize[J]. Applied Soil Ecology, 2016, 107: 324−333 doi: 10.1016/j.apsoil.2016.07.009 [18] CHEN L, LI K K, SHI W J, et al. Negative impacts of excessive nitrogen fertilization on the abundance and diversity of diazotrophs in black soil under maize monocropping[J]. Geoderma, 2021, 393: 114999 doi: 10.1016/j.geoderma.2021.114999 [19] 熊艺, 郑璐, 沈仁芳, 等. 缺氮胁迫对小麦根际土壤微生物群落结构特征的影响[J]. 土壤学报, 2022, 59(1): 218−230XIONG Y, ZHENG L, SHEN R F, et al. Effects of nitrogen deficiency on microbial community structure in rhizosphere soil of wheat[J]. Acta Pedologica Sinica, 2022, 59(1): 218−230 [20] 童依平, 李继云, 李振声. 不同小麦品种吸收利用氮素效率的差异及有关机理研究 Ⅰ. 吸收和利用效率对产量的影响[J]. 西北植物学报, 1999, 19(2): 270−277TONG Y P, LI J Y, LI Z S. Genotypic variations for nitrogen use efficiency in winter wheatⅠ. Effects of N uptake and utilization efficiency on grain yields[J]. Acta Botanica Boreali-Occidentalia Sinica, 1999, 19(2): 270−277 [21] SHI X L, CUI F, HAN X Y, et al. Comparative genomic and transcriptomic analyses uncover the molecular basis of high nitrogen-use efficiency in the wheat cultivar Kenong 9204[J]. Molecular Plant, 2022, 15(9): 1440−1456 doi: 10.1016/j.molp.2022.07.008 [22] 王晓婧, 代兴龙, 马鑫, 等. 不同小麦品种产量和氮素吸收利用的差异[J]. 麦类作物学报, 2017, 37(8): 1065−1071 doi: 10.7606/j.issn.1009-1041.2017.08.09WANG X J, DAI X L, MA X, et al. Differences of grain yield, nitrogen uptake and utilization efficiency of different wheat varieties[J]. Journal of Triticeae Crops, 2017, 37(8): 1065−1071 doi: 10.7606/j.issn.1009-1041.2017.08.09 [23] LIU J J, ZHANG Q, MENG D Y, et al. QMrl-7B enhances root system, biomass, nitrogen accumulation and yield in bread wheat[J]. Plants, 2021, 10(4): 764 doi: 10.3390/plants10040764 [24] WANG Y X, WANG C N, GU Y Z, et al. The variability of bacterial communities in both the endosphere and ectosphere of different niches in Chinese chives (Allium tuberosum)[J]. PLoS One, 2020, 15(1): e0227671 doi: 10.1371/journal.pone.0227671 [25] BOLYEN E, RIDEOUT J R, DILLON M R, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2[J]. Nature Biotechnology, 2019, 37(8): 852−857 doi: 10.1038/s41587-019-0209-9 [26] GLÖCKNER F O, YILMAZ P, QUAST C, et al. 25 years of serving the community with ribosomal RNA gene reference databases and tools[J]. Journal of Biotechnology, 2017, 261: 169−176 doi: 10.1016/j.jbiotec.2017.06.1198 [27] GLICK B R. Plant growth-promoting bacteria: mechanisms and applications[J]. Scientifica, 2012, 2012: 963401 [28] CASTRILLO G, TEIXEIRA P J P L, PAREDES S H, et al. Root microbiota drive direct integration of phosphate stress and immunity[J]. Nature, 2017, 543(7646): 513−518 doi: 10.1038/nature21417 [29] LIU H W, LI J Y, CARVALHAIS L C, et al. Evidence for the plant recruitment of beneficial microbes to suppress soil-borne pathogens[J]. New Phytologist, 2021, 229(5): 2873−2885 doi: 10.1111/nph.17057 [30] YIN C T, MUETH N, HULBERT S, et al. Bacterial communities on wheat grown under long-term conventional tillage and no-till in the Pacific Northwest of the United States[J]. Phytobiomes Journal, 2017, 1(2): 83−90 doi: 10.1094/PBIOMES-09-16-0008-R [31] WOLIŃSKA A, KUŹNIAR A, GAŁĄZKA A. Biodiversity in the rhizosphere of selected winter wheat (Triticum aestivum L. ) cultivars—genetic and catabolic fingerprinting[J]. Agronomy, 2020, 10(7): 953 doi: 10.3390/agronomy10070953 [32] SIMONIN M, DASILVA C, TERZI V, et al. Influence of plant genotype and soil on the wheat rhizosphere microbiome: evidences for a core microbiome across eight African and European soils[J]. FEMS Microbiology Ecology, 2020, 96(6): fiaa067 doi: 10.1093/femsec/fiaa067 [33] ZHANG J Y, LIU Y X, ZHANG N, et al. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice[J]. Nature Biotechnology, 2019, 37(6): 676−684 doi: 10.1038/s41587-019-0104-4 [34] SALEEM M, HU J E, JOUSSET A. More than the sum of its parts: microbiome biodiversity as a driver of plant growth and soil health[J]. Annual Review of Ecology, Evolution, and Systematics, 2019, 50: 145−168 doi: 10.1146/annurev-ecolsys-110617-062605 [35] EDWARDS J, JOHNSON C, SANTOS-MEDELLÍN C, et al. Structure, variation, and assembly of the root-associated microbiomes of rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(8): E911–E920 [36] WILLIAMS A, DE VRIES F T. Plant root exudation under drought: implications for ecosystem functioning[J]. New Phytologist, 2020, 225(5): 1899−1905 doi: 10.1111/nph.16223 [37] XIE H T, CHEN Z M, FENG X X, et al. L-theanine exuded from Camellia sinensis roots regulates element cycling in soil by shaping the rhizosphere microbiome assembly[J]. Science of the Total Environment, 2022, 837: 155801 doi: 10.1016/j.scitotenv.2022.155801 [38] 付博阳, 张钧浩, 杨明晓, 等. 追氮时期对强筋小麦根际土壤微生物群落结构的影响[J]. 河北农业大学学报, 2022, 45(3): 1−8FU B Y, ZHANG J H, YANG M X, et al. Effects of nitrogen topdressing timing on microbial community structure in strong gluten wheat rhizosphere soil[J]. Journal of Agricultural University of Hebei, 2022, 45(3): 1−8 [39] WARD L M, HEMP J, SHIH P M, et al. Evolution of phototrophy in the Chloroflexi phylum driven by horizontal gene transfer[J]. Frontiers in Microbiology, 2018, 9: 260 doi: 10.3389/fmicb.2018.00260 [40] 鲜文东, 张潇橦, 李文均. 绿弯菌的研究现状及展望[J]. 微生物学报, 2020, 60(9): 1801−1820 doi: 10.13343/j.cnki.wsxb.20200463XIAN W D, ZHANG X T, LI W J. Research status and prospect on bacterial phylum Chloroflexi[J]. Acta Microbiologica Sinica, 2020, 60(9): 1801−1820 doi: 10.13343/j.cnki.wsxb.20200463 [41] SPIECK E, SPOHN M, WENDT K, et al. Extremophilic nitrite-oxidizing Chloroflexi from Yellowstone hot springs[J]. The ISME Journal, 2020, 14(2): 364−379 doi: 10.1038/s41396-019-0530-9 [42] ANDREOTE F D, CARNEIRO R T, SALLES J F, et al. Culture-independent assessment of rhizobiales-related alphaproteobacteria and the diversity of Methylobacterium in the rhizosphere and rhizoplane of transgenic Eucalyptus[J]. Microbial Ecology, 2009, 57(1): 82−93 doi: 10.1007/s00248-008-9405-8 [43] NGOM M, OSHONE R, DIAGNE N, et al. Tolerance to environmental stress by the nitrogen-fixing actinobacterium Frankia and its role in actinorhizal plants adaptation[J]. Symbiosis, 2016, 70(1): 17−29 [44] 宁楚涵, 李文彬, 刘润进. 植物共生放线菌研究进展[J]. 生态学杂志, 2019, 38(1): 256−266 doi: 10.13292/j.1000-4890.201901.001NING C H, LI W B, LIU R J. Research advances in plant symbiotic actinomyces[J]. Chinese Journal of Ecology, 2019, 38(1): 256−266 doi: 10.13292/j.1000-4890.201901.001 [45] XUN W B, LIU Y P, LI W, et al. Specialized metabolic functions of keystone taxa sustain soil microbiome stability[J]. Microbiome, 2021, 9(1): 35 doi: 10.1186/s40168-020-00985-9 [46] WILLIAMS K P, SOBRAL B W, DICKERMAN A W. A robust species tree for the Alphaproteobacteria[J]. Journal of Bacteriology, 2007, 189(13): 4578−4586 doi: 10.1128/JB.00269-07 [47] MENDES R, GARBEVA P, RAAIJMAKERS J M. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms[J]. FEMS Microbiology Reviews, 2013, 37(5): 634−663 doi: 10.1111/1574-6976.12028 [48] UPADHYAY S K, SINGH J S, SAXENA A K, et al. Impact of PGPR inoculation on growth and antioxidant status of wheat under saline conditions[J]. Plant Biology, 2012, 14(4): 605−611 doi: 10.1111/j.1438-8677.2011.00533.x [49] 高金会, 张国良, 付卫东, 等. 基于宏基因组测序解析长刺蒺藜草入侵对根际土壤氮循环的影响[J]. 植物保护学报, 2022, 49(5): 1349−1357 doi: 10.13802/j.cnki.zwbhxb.2022.2022843GAO J H, ZHANG G L, FU W D, et al. Effects of spiny burr grass Cenchrus longispinus invasion on rhizosphere nitrogen cycle based on metagenome sequencing[J]. Journal of Plant Protection, 2022, 49(5): 1349−1357 doi: 10.13802/j.cnki.zwbhxb.2022.2022843 -





 下载:
下载: